1. Introduction¶
biobeam is an open software platform to rigorously simulate the image-formation process of fluorescent light microscopes, especially deep inside scattering biological tissues.
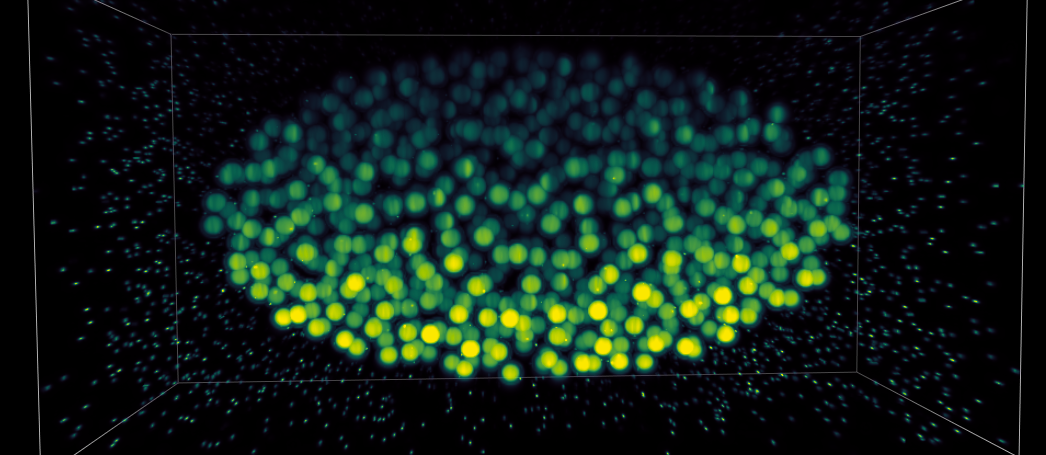
Synthetic embryo computationally imaged using a biobeam-implemented light sheet microscope.
For this biobeam integrates highly parallelized graphics processor units into a flexible computational pipe-lines starting with laser illumination of a specimen, and ending with partially coherent images, videos and 3d datasets, as recorded by a fluorescence camera.
biobeam includes modules for scalar and vectorial PSF calculations, fast GPU-acclerated beam propagation of arbitrary light fields through a given refractive index map, and the creation of simulated 3d image datasets from ground truth while taking for the whole image formation process in light-sheet microscopy into account.
The API is designed to be extremly simple and easy to use. See the following short demonstrations of the mentioned modules:
Scalar and vectorial PSF calculations
The following snippet calculates the 3d PSF of a gaussian beam with NA = 0.8 (both scalar and vectorial):
import biobeam
# return the intensity
u = m.biobeam.focus_field_beam(shape = (256,)*3,units = (.1,)*3,
NA = 0.8, n0 = 1.33)
# return all the complex vector components
u, ex,ey,ez = m.biobeam.focus_field_beam(shape = (256,)*3,units = (.1,)*3,
NA = 0.8, n0 = 1.33, return_all_fields = True)
Fast beam propagation light fields
The following snippet propagates different input fields through a given refractive index distribution:
import biobeam
# define the refractive index distribution
dn = ...
# set up the simulation object
m = Bpm3d(dn = dn, units = (0.1,)*3, lam = 0.5)
# propagate a plane wave and return the complex 3d field
u = m.propagate()
# propagate a bessel beam and return the 3d intensity
u = m.propagate(u0 = m.u0_beam(NA = (.4,.42)), return_comp = "intens")
Image formation process simulation in light-sheet microscopy
The following snippet simulated what a cylindrical light-sheet microscope (SPIM) with illumination NA=0.1 and detection NA = 0.6 would see of a given signal subject to a refractive index distribution at relative axial position z = 20um:
from biobeam import SimLSM_Cylindrical
#create some input data, r.i and labeled density
dn, signal = generate_some_refractive_volume_and_label()
#create a microscope simulator
m = SimLSM_Cylindrical(dn = dn, signal = signal,
NA_illum= .1, NA_detect=.6,
size = size, n0 = 1.33)
#simulate the image at relative axial position 20um
image = m.simulate_image_z(cz=20)
